Terminology
Before we start, there is some terminology that you'll need to know. Firstly, an enzyme is a protein that acts as a catalyst in biochemical reactions. They are globular proteins that exist in defined, 3D conformations. In order to catalyse a reaction, an enzyme must bind with a substance, called a substrate, to form an enzyme substrate complex. The substrate binds onto the enzyme's active site.
Some enzymes have other molecules, known as prosthetic groups, attached to them. Prosthetic groups are not part of the enzyme's polypeptide chain but are permanently attached to the enzyme. Prosthetic groups can be metal ions or other types of organic compounds. An example is the iron containing groups which are attached to the haemoglobin molecule.
Some enzymes, called allosteric enzymes, have an allosteric site. An allosteric site is the part of the enzyme molecule which binds with a non substrate molecule causing a conformational change. This change causes the enzyme's affinity for its substrate to change.
Isoenzymes are individual members of a set of enzymes which differ in amino acid composition but which can catalyse the same reaction. Isoenzymes differ in substrate affinity and maximum rates of enzyme-substrate interaction. They allow the 'fine tuning' of metabolism to meet the needs of a particular tissue.
How do catalysts work?
A catalyst is a substance that increases the rate of a chemical reaction without itself undergoing any permanent change. Catalysts do this by lowering the activation energy requirement of a reaction, causing it to occur faster. Catalysts work with equilibrium reactions (reversible reactions) and equilibrium and the enzyme itself are not changed.
 | |
| An example of how an enzyme affects Activation Energy. Source: http://commons.wikimedia.org/wiki/File%3AActivation2.png Please see http://creativecommons.org/licenses/by-sa/3.0/ if you'd like to use this diagram. |
Models of Enzyme-Substrate Interactions
Currently, there are two main models which explain how the enzyme molecule interacts with the substrate molecule.
The Lock and Key Model:
In the lock and key model, the enzyme's active site is the 'lock' and the substrate is the 'key'. The 3D structure of the lock fits exactly with the structure of the substrate. Because enzymes are specific only the right substrate can fit with the right enzyme. However, this model is too rigid as it doesn't take into consideration the effect of allosteric molecules.
The Induced Fit Model:
This model says that the initial reaction between enzyme and substrate is weak but causes a conformational change in the enzyme which strengthens the bonding between the two molecules. This allows the catalytic site to get closer to the substrate bonds which are to be altered. The configuration of the active site is altered and this positions the reactive groups of the enzyme in a position which is optimal for the catalytic reaction. Once this bonding has occurred there are 4 possible ways for catalysis to take place:
- Catalysis by bond strain: Structural rearrangements caused by the bonding of the enzyme and substrate strain the substrate bonds causing them to be more reactive. This lowers the energy needed for the reaction to occur.
- Catalysis by Proximity and Orientation: When substrate molecules are free in solution they move around and collide. This lowers the probability that the collision will occur at the angle required for the reaction to proceed. Enzymes align the chemical groups and hold them close together making the reaction more likely to occur. The enzyme holds the substrate in an orientation appropriate for a collision.
- Catalysis involving proton donors (acids) and acceptors (bases): The acidic or basic side chains of the enzyme's amino acids form the active site and transfer protons to or from the substrate. This destabilises the covalent bonds in the substrate, making the substrate more reactive.
- Covalent Catalysis: In this type of catalysis, the substrate is oriented to the active sites of the enzyme in a way that causes a covalent intermediate to form between the enzyme and the substrate. This helps to reduce the amount of energy required in later stages of the reaction.
Organophosphates:
They bind with reactive serine amino acids to form an inactive complex. They affect serine proteases (enzymes which cleave peptide bonds in some proteins). An example is acetylcholinesterase which is involved in the transmission of nerve impulses. If acetlycholinesterase no longer functions, continued depolarisation of nerves occur causing symptoms such as: constricted pupils, vomiting, diarrhoea, muscle tremors and weakness progressing to paralysis and death.
Heavy Metals:
They bind to sulphur so that any enzyme with a cysteine covalent intermediate will be inactivated by metals such as: silver, mercury, lead and copper. An example is lead poisoning which interrupts the synthesis of haemoglobin.
How we can use enzymes to diagnose disease
When the cells of a tissue become damaged or die, their contents spill into the extra cellular fluid. Their contents eventually end up in blood plasma or in lymph. By examining the enzyme activities in blood the location and extent of the disease can be identified. This is because tissues often have enzymes which are specific to them. For example, if a person has suffered from a heart attack the levels of creatinine kinase is likely to be elevated.
Slave and Regulatory Enzymes
Regulatory enzymes are those whose activities are increased or decreased by specific mechanisms. Slave enzymes (also known as Michaelis-Menten enzymes) are those whose activities are controlled by their environment.
Factors which affect all enzymes
The following factors affect all enzymes:
- pH: The pH alters the charge of a protein, the electrostatic bonds which bind the protein together (and hence its tertiary structure), and substrate binding because of changes in geometry and charge of the active site.
- Temperature: A small change in temperature will increase the reactions rate of the enzyme. This is because as the temperature increases so does the kinetic energy of both the enzyme and substrate resulting in more frequent collisions between the two. This increases the chance that the correct substrate will bind with its corresponding enzyme and provides enough energy to undergo a reaction. However, if the temperature gets too high denaturation of the enzyme may result.
- Substrate concentration: the more substrate is added to a solution containing the correct enzyme, the faster the reaction will occur until the enzymes are saturated.
- inhibitors
- enzyme concentrations
Below is a typical substrate saturation curve for a Michaelis-Menten enzyme:
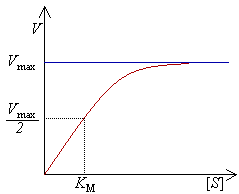 |
| source: http://commons.wikimedia.org/wiki/File%3AMichaelis-Menten.png please see http://creativecommons.org/licenses/by-sa/3.0/deed.en if you'd like to use this diagram |
 |
| source: http://commons.wikimedia.org/wiki/File%3AMichaelis_menten_equation.jpg please see http://creativecommons.org/licenses/by-sa/3.0/deed.en if you'd like to use this picture |
Measuring the Activity of Enzymes
When we measure the activity of enzymes, the enzyme must be the only reactant that limits the reaction. In other words, all the other reactants must be present in an excess amount. The substrate concentration must also be in excess. Zero order kinetics must be present which means that at high substrate concentrations the reaction rate is independent of the substrate concentration. This is because adding more substrate will not increase the reaction rate (this is known as zero order with respect to substrate). Under conditions of zero order kinetics, the velocity is dependent on the amount of enzyme available. We can use this to determine the enzyme activity.
Enzyme activity can be measured by determining the rate of a substrate disappearing, the rate of appearance of a product or with the use of a spectrophotometer.
Enzyme Inhibition
There are many different substances which can inhibit the activity of enzymes. Examples include: substrate analogs, toxins, drugs and metal complexes. Enzyme inhibition is clinically important because many drugs work by inhibiting specific enzymes. For example, sulfa drugs inhibit the synthesis of folic acid in bacteria, causing them to die.
There are two main categories of enzyme inhibitors:
- Irreversible inhibitors: which cause covalent modification of the enzyme structure. These are considered poisons and are generally not used in treatment.
- Reversible inhibitors: of which there are three types:
- Competitive Inhibitors: the inhibitor resembles the normal substrate and competes with it for the same site. Inhibition is reversible by substrate. This causes Vmax to remain unchanged while Km is increased.
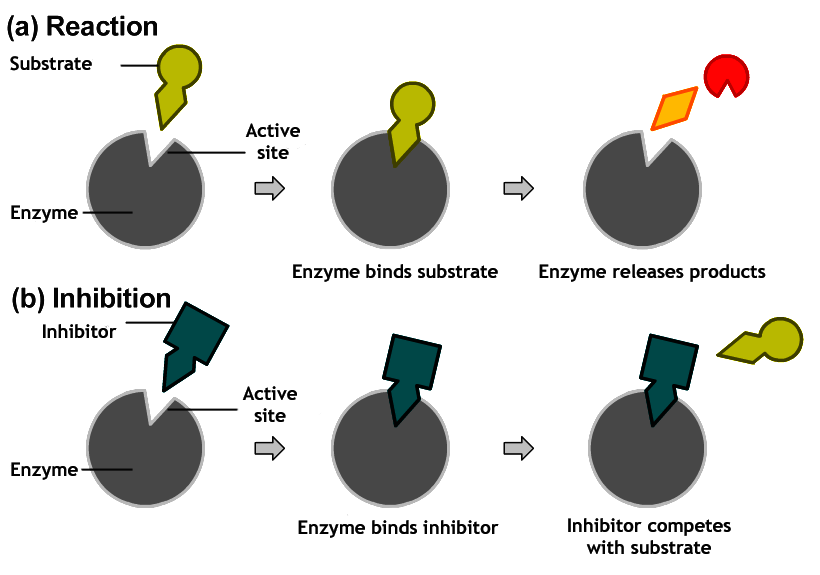 |
| How competitive inhibition works |
| Reaction scheme for competitive inhibition. E=enzyme; S=substrate; I = inhibitor; ES= enzyme substrate complex P= product; EI= Enzyme-inhibitor complex |
- Noncompetitive inhibitors: can bind to the enzyme before the substrate. The inhibitor binds to the enzyme or enzyme-substrate complex other than at the active site. This doesn't affect substrate binding but the enzyme-substrate-inhibitor complex can't form products. Inhibition cannot be reversed by the substrate. The Km doesn't change but Vmax is decreased.
| Reaction scheme for noncompetitive inhibition |
| Effect of noncompetitive inhibition on Vmax and Km. (1) = without inhibitor; (2)= with inhibitor |
- Uncompetitive inhibitors: can't bind before the substrate. The inhibitor only binds to the enzyme-substrate complex at areas other than the active site. Substrate binding modifies the enzyme structure, making an inhibitor-binding site available. Inhibition cannot be reversed by the substrate. Vmax and Km are decreased.
| Effect of uncompetitive inhibition on Km and Vmax. |
| Reaction scheme for uncompetitive inhibition |
Regulation of Enzyme Activity
The regulation of enzymes is important because it allows the cell to efficiently use its resources. The activity of regulatory enzymes can be altered in a number of ways:
- Rate of enzyme synthesis and degradation
- Proenzyme activiation: a rapid method of increasing enzyme activity but is not reversible. Proenzymes are synthesised in abundance and then stored in secretory granules and activated upon release from their storage sites.
- Compartmentalisation: enzymes are separated into areas where the availability of their substrate is limited.
- Modification of regulatory enzyme activity
- stimulation and inhibition by control proteins
- allosteric interactions
Allosteric enzymes have three important properties:
- They have multiple subunits; they have separate catalytic and regulatory subunits. Individual catalytic subunits each have their own active site.
- They exist in two forms:
- Active
- Inactive
- They display a characteristic 'S' shaped curve for a reaction rate vs substrate concentration graph.